Lobry-Rosso-Flandros (LRF) model for fitting thermal performance curves
Source:R/lrf_1991.R
lrf_1991.RdLobry-Rosso-Flandros (LRF) model for fitting thermal performance curves
Value
a numeric vector of rate values based on the temperatures and parameter values provided to the function
Details
Equation: $$rate= rmax \cdot \frac{(temp - t_{max}) \cdot (temp - t_{min})^2}{(t_{opt} - t_{min}) \cdot ((t_{opt} - t_{min}) \cdot (temp - t_{opt}) - (t_{opt} - t_{max}) \cdot (t_{opt} + t_{min} - 2 \cdot temp))}$$
Start values in get_start_vals are derived from the data.
Limits in get_lower_lims and get_upper_lims are derived from the data or based extreme values that are unlikely to occur in ecological settings.
References
Rosso, L., Lobry, J. R., & Flandrois, J. P. An unexpected correlation between cardinal temperatures of microbial growth highlighted by a new model. Journal of Theoretical Biology, 162(4), 447-463. (1993)
Examples
# load in ggplot
library(ggplot2)
library(nls.multstart)
# subset for the first TPC curve
data('chlorella_tpc')
d <- subset(chlorella_tpc, curve_id == 1)
# get start values and fit model
start_vals <- get_start_vals(d$temp, d$rate, model_name = 'lrf_1991')
# fit model
mod <- nls_multstart(rate~lrf_1991(temp = temp, rmax, topt, tmin, tmax),
data = d,
iter = c(3,3,3,3),
start_lower = start_vals - 10,
start_upper = start_vals + 10,
lower = get_lower_lims(d$temp, d$rate, model_name = 'lrf_1991'),
upper = get_upper_lims(d$temp, d$rate, model_name = 'lrf_1991'),
supp_errors = 'Y',
convergence_count = FALSE)
# look at model fit
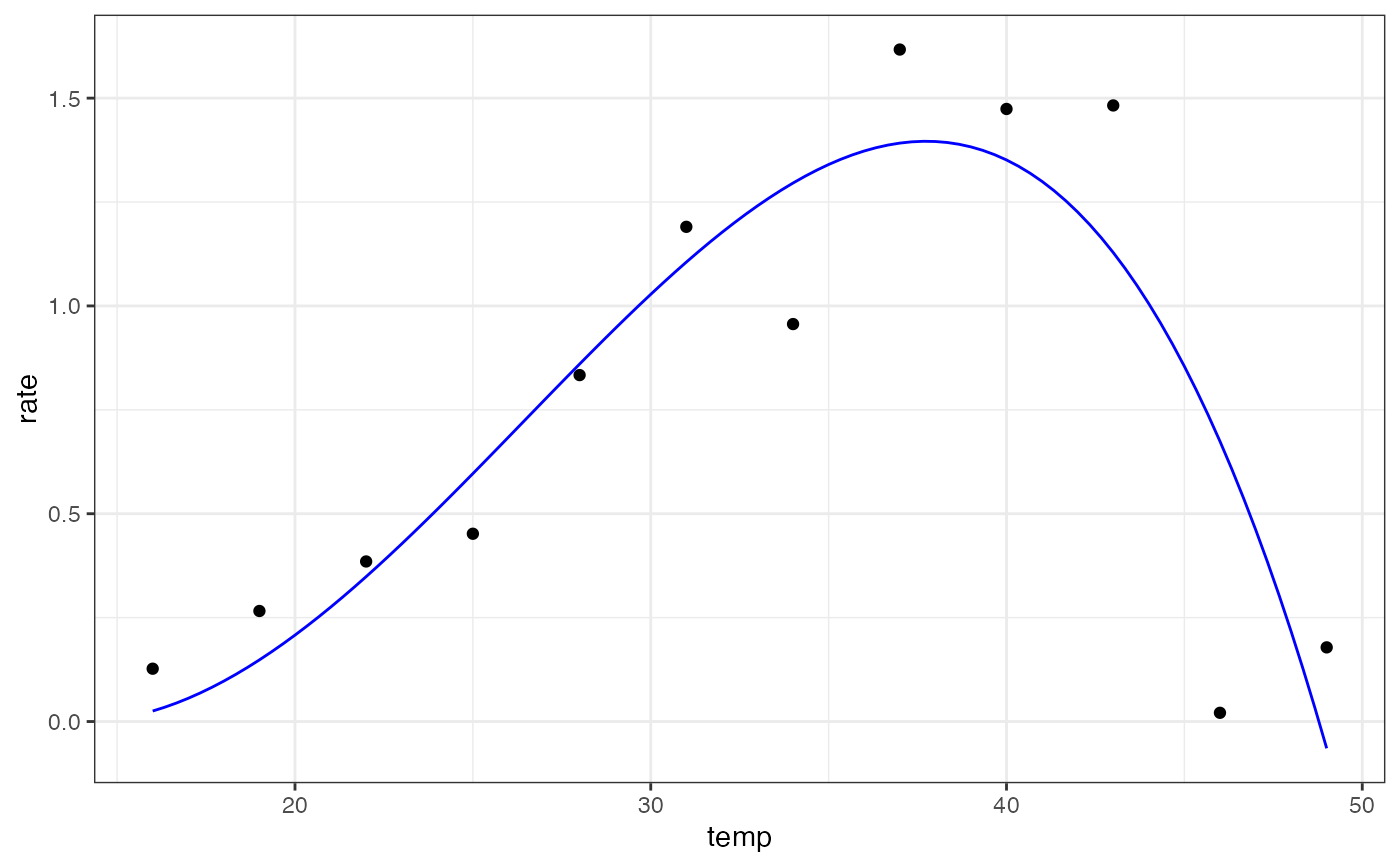
summary(mod)
#>
#> Formula: rate ~ lrf_1991(temp = temp, rmax, topt, tmin, tmax)
#>
#> Parameters:
#> Estimate Std. Error t value Pr(>|t|)
#> rmax 1.3962 0.1607 8.688 2.40e-05 ***
#> topt 37.7416 2.0042 18.831 6.54e-08 ***
#> tmin 14.0262 5.8327 2.405 0.0429 *
#> tmax 48.7822 0.9827 49.643 3.00e-11 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 0.3253 on 8 degrees of freedom
#>
#> Number of iterations to convergence: 46
#> Achieved convergence tolerance: 1.49e-08
#>
# get predictions
preds <- data.frame(temp = seq(min(d$temp), max(d$temp), length.out = 100))
preds <- broom::augment(mod, newdata = preds)
# plot
ggplot(preds) +
geom_point(aes(temp, rate), d) +
geom_line(aes(temp, .fitted), col = 'blue') +
theme_bw()